Python GEKKO ODE unexpected result
I’ve been trying to implement ODE models to model the insulin signaling pathway, as shown in <a href=”https://www.jbc.org/article/S0021-9258(19)48589-X/fulltext#seccestitle10″ rel=”noreferrer noopener nofollow”>this paper , Use python’s GEKKO
The implemented model variant is “Md3” with the following equations, constants, and initial values:
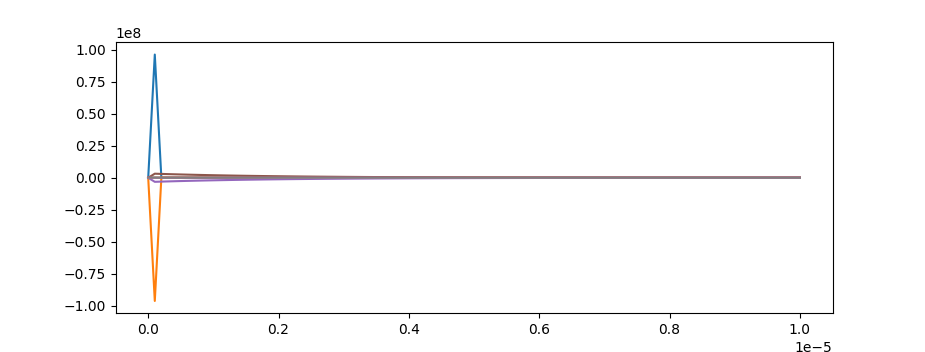
Even if this paper does not provide the result of the supplementary code, one would expect the result to be a curve, not the result produced by my code: 
I’VE CHECKED EQUATIONS AND CONSTANT VALUES, TRIED ADDING LOWER AND UPPER BOUNDS FOR VARIABLES, AND TRIED USING THE M.OPTIONS.IMODE AND M.OPTIONS.NODES PARAMETERS, BUT THESE DON’T SEEM TO HELP.
Any suggestions would be appreciated.
def insulin_pathway_CM(time_interval, insulin=0):
m = GEKKO()
t = np.linspace(0, time_interval-1, time_interval)
m.time = np.insert(t,1,[1e-5,1e-4,1e-3,1e-2,0.1])
# initialization of variables
ins = m.Param(value=insulin)
IR = m.Var(10)
IRp = m.Var()
IRS = m.Var(10)
IRSp = m.Var()
PKB = m.Var(10)
PKBp = m.Var()
GLUT4_C = m.Var(10)
GLUT4_M = m.Var()
# initialization of constants
k1aBasic = 1863.78
k1f = 38668.300000000003
k1b = 40229000
k2f = 401394
k2b = 36704300
k4f = 336782
k4b = 187399
k5Basic = 85530.899999999994
k5f = 11264.799999999999
k5b = 26389900
# equations
m.Equation(IR.dt() == k1b*IRp - (k1f*ins*1000*IR + k1aBasic*IR))
m.Equation(IRp.dt() == -k1b*IRp + k1f*ins*1000*IR + k1aBasic*IR)
m.Equation(IRS.dt() == k2b*IRSp - (k2f*IRp*IRS))
m.Equation(IRSp.dt() == -k2b*IRSp + k2f*IRp*IRS)
m.Equation(PKB.dt() == k4b*PKBp - k4f*IRSp*PKB)
m.Equation(PKBp.dt() == -k4b*PKBp + k4f*IRSp*PKB)
m.Equation(GLUT4_C.dt() == k5b*GLUT4_M - (k5f*PKBp*GLUT4_C + k5Basic*GLUT4_C))
m.Equation(GLUT4_M.dt() == -k5b*GLUT4_M + (k5f*PKBp*GLUT4_C + k5Basic*GLUT4_C))
m.options.IMODE = 7
m.options.OTOL = 1e-8
m.options.RTOL = 1e-8
m.options.NODES = 3
m.solve(disp=False)
# plotting
fig, axs = plt.subplots(4, 2, figsize=(20, 20))
axs[0, 0].plot(m.time, IR)
axs[0, 0].set_title('IR(t)')
axs[0, 1].plot(m.time, IRp)
axs[0, 1].set_title('IRp(t)')
axs[1, 0].plot(m.time, IRS, 'tab:orange')
axs[1, 0].set_title('IRS(t)')
axs[1, 1].plot(m.time, IRSp, 'tab:green')
axs[1, 1].set_title('IRSp(t)')
axs[2, 0].plot(m.time, PKB, 'tab:red')
axs[2, 0].set_title('PKB(t)')
axs[2, 1].plot(m.time, PKBp, 'tab:purple')
axs[2, 1].set_title('PKBp(t)')
axs[3, 0].plot(m.time, GLUT4_C, 'tab:olive')
axs[3, 0].set_title('GLUT4_C(t)')
axs[3, 1].plot(m.time, GLUT4_M, 'tab:cyan')
axs[3, 1].set_title('GLUT4_M(t)')
plt.figure()
plt.show()
return
time_interval = 60
insulin = 100
insulin_pathway_CM(time_interval, insulin)
Solution
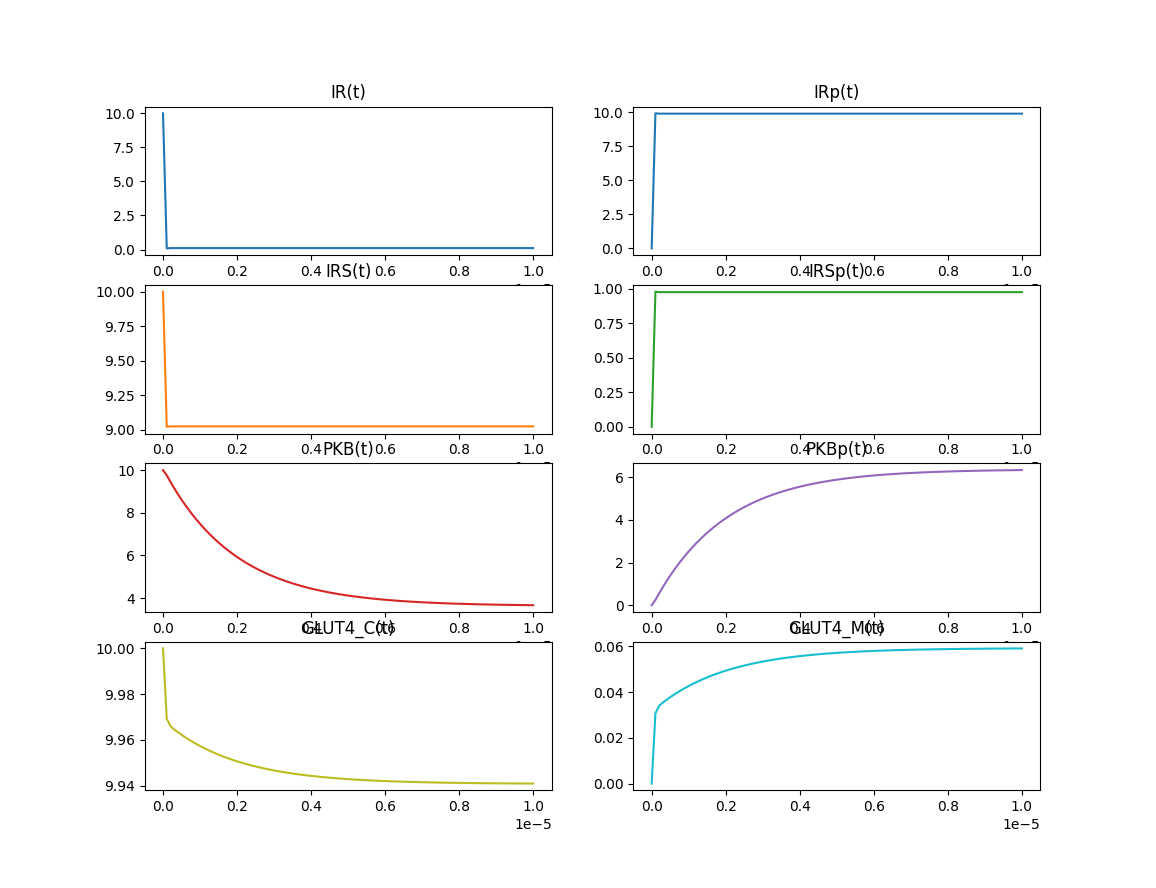
Lutz Lehmann is right. Derivative plots with end times of 1e-5 show that most of the action occurs over a short period of time.
Try to shorten the time span.
m.time = np.linspace(0,1e-5,100)
This shows rapid dynamics.
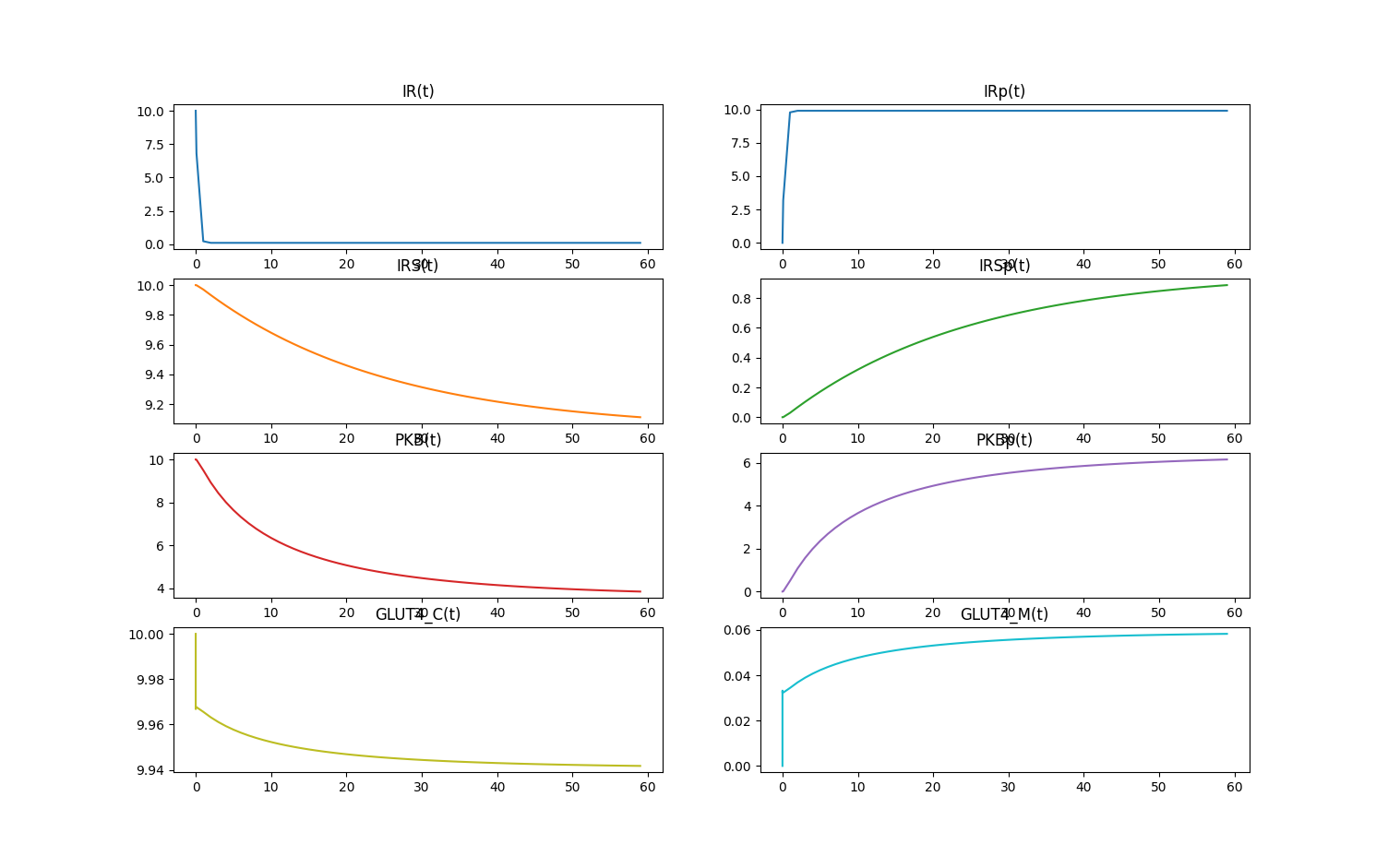
There may be errors in the equation, such as a unit problem (time in days or years?). Or the author forgot to include some type of volume factor for the patient, such as blood volume or volume.
# equations
V1 = 1e9 # hypothetical volume 1
V2 = 10 # hypothetical volume 2
m.Equation(V1*IR.dt() == k1b*IRp - (k1f*ins*1000*IR + k1aBasic*IR))
m.Equation(V1*IRp.dt() == -k1b*IRp + k1f*ins*1000*IR + k1aBasic*IR)
m.Equation(V1*IRS.dt() == k2b*IRSp - (k2f*IRp*IRS))
m.Equation(V1*IRSp.dt() == -k2b*IRSp + k2f*IRp*IRS)
m.Equation(V2*PKB.dt() == k4b*PKBp - k4f*IRSp*PKB)
m.Equation(V2*PKBp.dt() == -k4b*PKBp + k4f*IRSp*PKB)
m.Equation(V2*GLUT4_C.dt() == k5b*GLUT4_M - (k5f*PKBp*GLUT4_C + k5Basic*GLUT4_C))
m.Equation(V2*GLUT4_M.dt() == -k5b*GLUT4_M + (k5f*PKBp*GLUT4_C + k5Basic*GLUT4_C))
Here are more plausible dynamics on the original timescale (0-60 hours).
from gekko import GEKKO
import numpy as np
import matplotlib.pyplot as plt
def insulin_pathway_CM(time_interval, insulin=0):
m = GEKKO()
t = np.linspace(0, time_interval-1, time_interval)
m.time = np.insert(t,1,[1e-5,1e-4,1e-3,1e-2,0.1])
# initialization of variables
ins = m.Param(value=insulin)
IR = m.Var(10)
IRp = m.Var()
IRS = m.Var(10)
IRSp = m.Var()
PKB = m.Var(10)
PKBp = m.Var()
GLUT4_C = m.Var(10)
GLUT4_M = m.Var()
# initialization of constants
k1aBasic = 1863.78
k1f = 38668.300000000003
k1b = 40229000
k2f = 401394
k2b = 36704300
k4f = 336782
k4b = 187399
k5Basic = 85530.899999999994
k5f = 11264.799999999999
k5b = 26389900
# calculate derivatives
d = m.Array(m.Var,8)
m.Equation(d[0] == k1b*IRp - (k1f*ins*1000*IR + k1aBasic*IR))
m.Equation(d[1] == -k1b*IRp + k1f*ins*1000*IR + k1aBasic*IR)
m.Equation(d[2] == k2b*IRSp - (k2f*IRp*IRS))
m.Equation(d[3] == -k2b*IRSp + k2f*IRp*IRS)
m.Equation(d[4] == k4b*PKBp - k4f*IRSp*PKB)
m.Equation(d[5] == -k4b*PKBp + k4f*IRSp*PKB)
m.Equation(d[6] == k5b*GLUT4_M - (k5f*PKBp*GLUT4_C + k5Basic*GLUT4_C))
m.Equation(d[7] == -k5b*GLUT4_M + (k5f*PKBp*GLUT4_C + k5Basic*GLUT4_C))
# equations
V1 = 1e9 # hypothetical volume 1
V2 = 10 # hypothetical volume 2
m.Equation(V1*IR.dt() == k1b*IRp - (k1f*ins*1000*IR + k1aBasic*IR))
m.Equation(V1*IRp.dt() == -k1b*IRp + k1f*ins*1000*IR + k1aBasic*IR)
m.Equation(V1*IRS.dt() == k2b*IRSp - (k2f*IRp*IRS))
m.Equation(V1*IRSp.dt() == -k2b*IRSp + k2f*IRp*IRS)
m.Equation(V2*PKB.dt() == k4b*PKBp - k4f*IRSp*PKB)
m.Equation(V2*PKBp.dt() == -k4b*PKBp + k4f*IRSp*PKB)
m.Equation(V2*GLUT4_C.dt() == k5b*GLUT4_M - (k5f*PKBp*GLUT4_C + k5Basic*GLUT4_C))
m.Equation(V2*GLUT4_M.dt() == -k5b*GLUT4_M + (k5f*PKBp*GLUT4_C + k5Basic*GLUT4_C))
m.options.IMODE = 4
m.options.OTOL = 1e-8
m.options.RTOL = 1e-8
m.options.NODES = 3
m.solve(disp=True)
# plotting
fig, axs = plt.subplots(4, 2, figsize=(20, 20))
axs[0, 0].plot(m.time, IR)
axs[0, 0].set_title('IR(t)')
axs[0, 1].plot(m.time, IRp)
axs[0, 1].set_title('IRp(t)')
axs[1, 0].plot(m.time, IRS, 'tab:orange')
axs[1, 0].set_title('IRS(t)')
axs[1, 1].plot(m.time, IRSp, 'tab:green')
axs[1, 1].set_title('IRSp(t)')
axs[2, 0].plot(m.time, PKB, 'tab:red')
axs[2, 0].set_title('PKB(t)')
axs[2, 1].plot(m.time, PKBp, 'tab:purple')
axs[2, 1].set_title('PKBp(t)')
axs[3, 0].plot(m.time, GLUT4_C, 'tab:olive')
axs[3, 0].set_title('GLUT4_C(t)')
axs[3, 1].plot(m.time, GLUT4_M, 'tab:cyan')
axs[3, 1].set_title('GLUT4_M(t)')
plt.figure()
for i in range(8):
plt.plot(m.time,d[i].value)
plt.show()
return
time_interval = 60
insulin = 100
insulin_pathway_CM(time_interval, insulin)
As a reference, here’s a simplified blood glucose response model (similar to the Bergman model) for everyone to quote. Richard Bergman and Claudio Cobelli proposed a minimal model describing blood glucose and insulin dynamics in 1979.