Create faceted matplotlib/seaborn plots using indicator variables instead of single columns
Seaborn is ideal for creating polygon charts based on categorical variables that encode the categories of each polygon. However, this assumes that your categories are mutually exclusive. Is it possible to create a Seaborn FacetGrid (or similar) based on a set of metric variables?
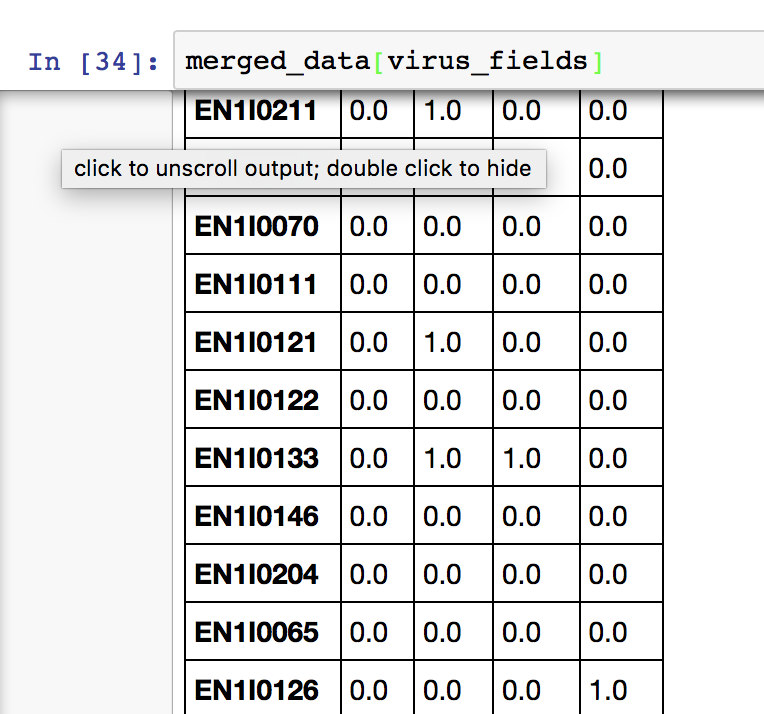
As a concrete example, consider comparing patients infected with one or more viruses and plotting attributes of interest by virus. A patient may carry more than one virus, so it is not possible to create a virus column to create a grid. However, you can create a set of indicator variables, one for each virus, to label the virus for each patient. There seems to be no way to pass a set of indication variables to any Seaborn function to do this.
I
can’t imagine I’m the first to come across this, so I’d like advice on how to do this without having to hand-code in Matplotlib.
Solution
I don’t see how to use FacetGrid to do this, probably because this is not faceting the data, as the data record may appear multiple times or only once in the graph. One of the standard tricks for a set of bitfields is to read them as binary so you can see every combination of bits. This is unambiguous, but can get confusing:
import pandas as pd
import seaborn as sns
from numpy.random import random, randint
from numpy import concatenate
import matplotlib.pyplot as plt
# Dummy data
vdata = pd. DataFrame(concatenate((randint(2, size=(32,4)), random(size=(32,2))), axis=1))
vdata.columns=['Species','v1','v2','v3','x','y']
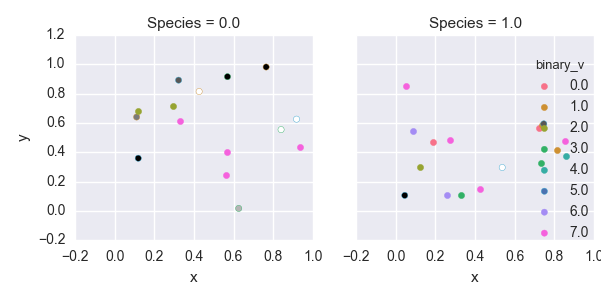
binary_v = vdata.v1 + vdata.v2*2 + vdata.v3*4
# Making a binary number out of the "virusX?" fields
pd.concat((vdata, binary_v), axis=1)
vdata = pd.concat((vdata, binary_v), axis=1)
vdata.columns=['Species','v1','v2','v3','x','y','binary_v']
# Plotting group membership by row
#g = sns. FacetGrid(vdata, col="Species", row='binary_v')
#g.map(plt.scatter, "x", "y")
#g.add_legend()
#plt.savefig('multiple_facet_binary_row') # Unreadably big.
h = sns. FacetGrid(vdata, col="Species", hue="binary_v")
h.map(plt.scatter, "x","y")
h.add_legend()
plt.savefig('multiple_facet_binary_hue')
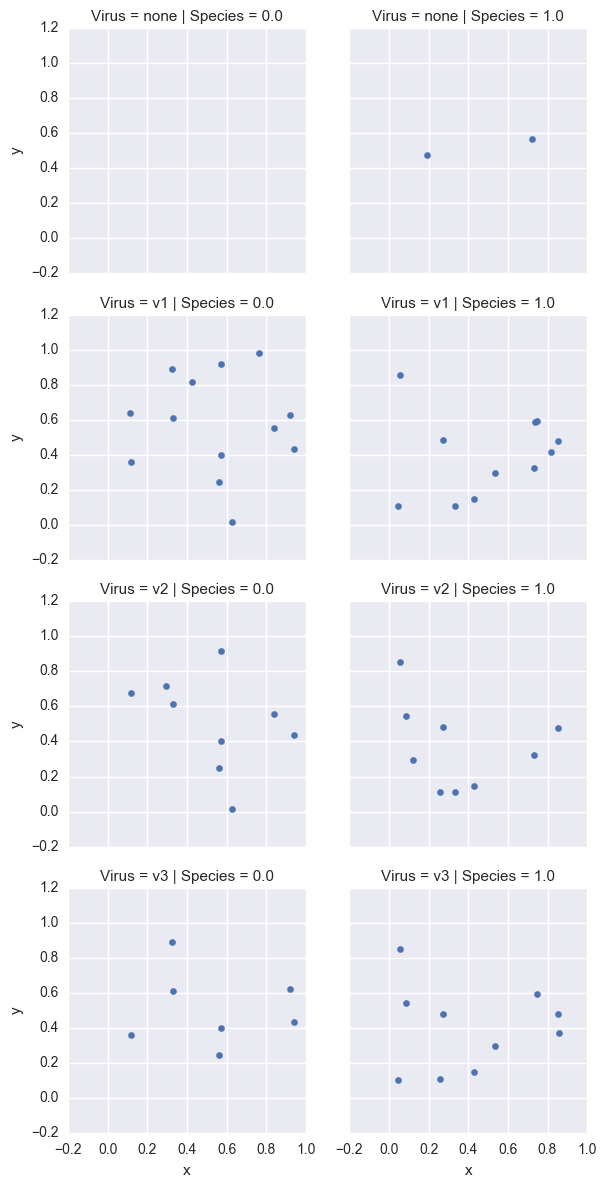
If you have too many metrics to handle combinatorial explosions, explicitly make the new subset work:
# Nope, need to pull out subsets:
bdata = vdata[vdata.v1 + vdata.v2 + vdata.v3 ==0.]
assert(len(bdata) > 0) # ... catch...
bdata['Virus'] = pd. Series(['none']*len(bdata), index=bdata.index)
for i in ['v1','v2','v3']:
on = vdata[vdata[i]==1.]
on['Virus'] = pd. Series([i]*len(on), index=on.index)
bdata = bdata.append(on)
j = sns. FacetGrid(bdata, col='Species', row='Virus')
j.map(plt.scatter, 'x', 'y')
j.add_legend()
j.savefig('multiple_facet_refish')