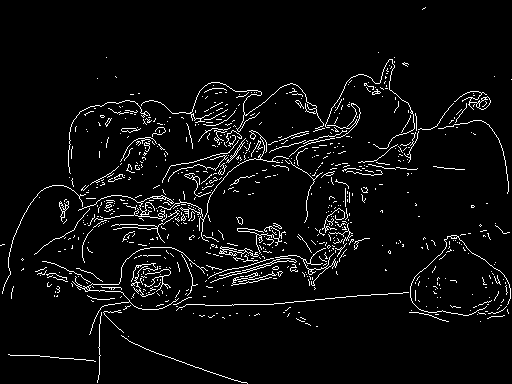What is the OpenCv equivalent of this Matlab code for Sobel edge detection?
This is the Matlab code I want to replicate in OpenCv
e[~, threshold] = edge(I, 'sobel');
fudgeFactor = .5;
BWs = edge(I,'sobel', threshold * fudgeFactor);
figure, imshow(BWs), title('binary gradient mask');
Here is my test image:
I’ve tried something similar
blurred_gray = cv2. GaussianBlur(gray_image,(3,3),0)
sobelx = cv2. Sobel(blurred_gray,cv2. CV_8U,1,0,ksize=3)
sobely = cv2. Sobel(blurred_gray,cv2. CV_8U,0,1,ksize=3)[2]
The output I get is:
I
tried adding sobelx and sobely because I read that they are partial derivatives, but the resulting image looks the same as above and changing ksize doesn’t help.
This is the output I need:
Can anyone tell me what I did wrong and what should I do to get the same result image?
Solution
The MATLAB implementation of Sobel edge detection is not visible, so we can only guess what exactly happened. The only hint we get from the documentation on edge is when using the 'sobel' option
Finds edges at those points where the gradient of the image I is
maximum, using the Sobel approximation to the derivative.
Although not stated, taking the maximum value of the gradient is much more complicated than simply taking the local maximum value in the image. Instead, we want to find the local maximum value with respect to the direction of the gradient. Unfortunately, the actual code that MATLAB uses for this is hidden.
Looking at the code available in Edge, they seem to use 4*mean(magnitude) as a threshold in the refinement operation, so I use it in conjunction with your gummy factor. orientated_non_max_suppression function is far from optimal, but I wrote it to improve readability rather than performance.
import cv2
import numpy as np
import scipy.ndimage.filters
gray_image = cv2.imread('cell.png', cv2. IMREAD_GRAYSCALE).astype(dtype=np.float32)
def orientated_non_max_suppression(mag, ang):
ang_quant = np.round(ang / (np.pi/4)) % 4
winE = np.array([[0, 0, 0],
[1, 1, 1],
[0, 0, 0]])
winSE = np.array([[1, 0, 0],
[0, 1, 0],
[0, 0, 1]])
winS = np.array([[0, 1, 0],
[0, 1, 0],
[0, 1, 0]])
winSW = np.array([[0, 0, 1],
[0, 1, 0],
[1, 0, 0]])
magE = non_max_suppression(mag, winE)
magSE = non_max_suppression(mag, winSE)
magS = non_max_suppression(mag, winS)
magSW = non_max_suppression(mag, winSW)
mag[ang_quant == 0] = magE[ang_quant == 0]
mag[ang_quant == 1] = magSE[ang_quant == 1]
mag[ang_quant == 2] = magS[ang_quant == 2]
mag[ang_quant == 3] = magSW[ang_quant == 3]
return mag
def non_max_suppression(data, win):
data_max = scipy.ndimage.filters.maximum_filter(data, footprint=win, mode='constant')
data_max[data != data_max] = 0
return data_max
# compute sobel response
sobelx = cv2. Sobel(gray_image, cv2. CV_32F, 1, 0, ksize=3)
sobely = cv2. Sobel(gray_image, cv2. CV_32F, 0, 1, ksize=3)
mag = np.hypot(sobelx, sobely)
ang = np.arctan2(sobely, sobelx)
# threshold
fudgefactor = 0.5
threshold = 4 * fudgefactor * np.mean(mag)
mag[mag < threshold] = 0
# non-maximal suppression
mag = orientated_non_max_suppression(mag, ang)
# alternative but doesn't consider gradient direction
# mag = skimage.morphology.thin(mag.astype(np.bool)).astype(np.float32)
# create mask
mag[mag > 0] = 255
mag = mag.astype(np.uint8)
Cell results
python
MATLAB
Results on MATLAB’s peppers .png (built-in).
python
MATLAB
The MATLAB implementation has to use something different, but it looks close.